How To Filter Out Na In R
R is.na Function Example (remove, supercede, count, if else, is not NA)
Well, I estimate it goes without saying that NA values decrease the quality of our information.
Fortunately, the R programming linguistic communication provides us with a role that helps usa to deal with such missing information: the is.na function.
In the following article, I'm going to explicate what the part does and how the function tin can be applied in exercise.
Let'south swoop in…
The is.na Function in R (Basics)
Before nosotros can first, let's create some example data in R (or R Studio).
ready . seed ( 11991 ) # Fix seed N <- 1000 # Sample size x_num <- round(rnorm(N, 0, v ) ) # Numeric x_fac <- as . factor (round(runif(North, 0, 3 ) ) ) # Factor x_cha <- sample(letters, N, supplant = TRUE ) # Character x_num[rbinom(North, 1, 0.2 ) == ane ] <- NA # 20% missings x_fac[rbinom(N, 1, 0.3 ) == 1 ] <- NA # 30% missings x_cha[rbinom(N, ane, 0.05 ) == ane ] <- NA # 5% missings information <- data. frame (x_num, x_fac, x_cha, # Create information frame stringsAsFactors = FALSE ) head(data) # First rows of information
set.seed(11991) # Set seed N <- grand # Sample size x_num <- round(rnorm(North, 0, 5)) # Numeric x_fac <- as.gene(round(runif(N, 0, iii))) # Factor x_cha <- sample(messages, N, replace = TRUE) # Character x_num[rbinom(N, one, 0.ii) == 1] <- NA # 20% missings x_fac[rbinom(N, 1, 0.three) == 1] <- NA # 30% missings x_cha[rbinom(Due north, ane, 0.05) == 1] <- NA # five% missings data <- data.frame(x_num, x_fac, x_cha, # Create data frame stringsAsFactors = FALSE) head(information) # Get-go rows of data
Our data consists of three columns, each of them with a different class: numeric, factor, and graphic symbol. This is how the first half dozen lines of our information look like:
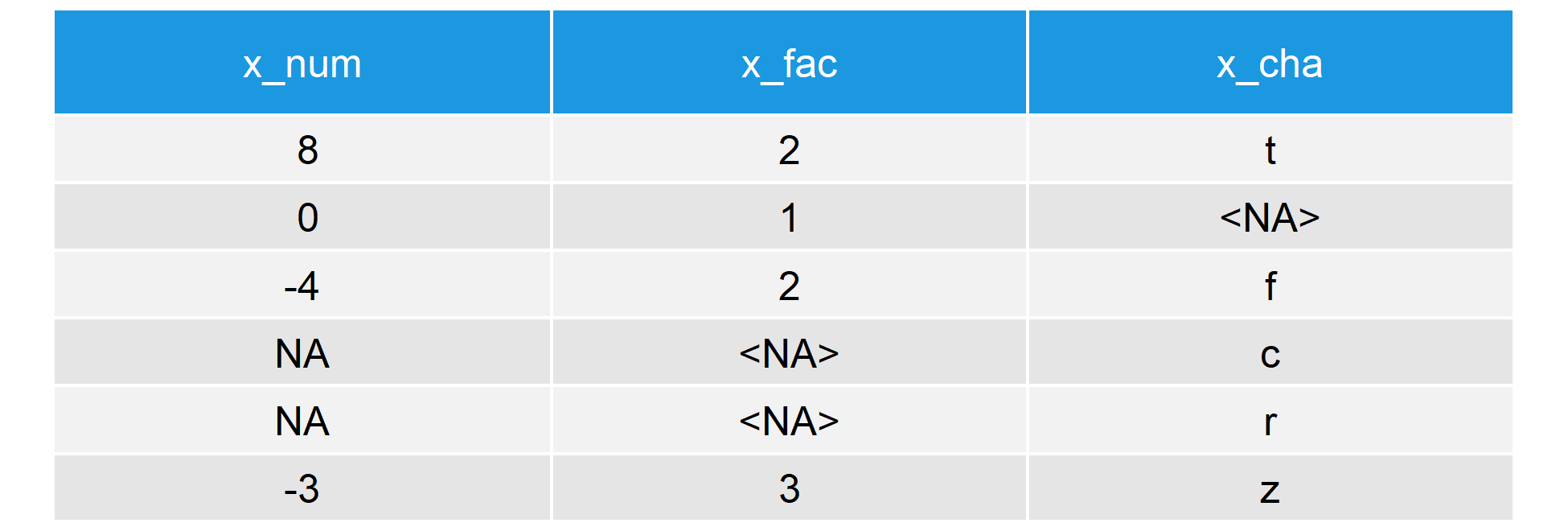
Table ane: Example Data for the is.na R Function (Showtime six Rows)
Let'southward employ the is.na part to our whole information set:
is . na (data) # x_num x_fac x_cha # [1,] False Fake Imitation # [two,] FALSE FALSE Truthful # [3,] False FALSE Fake # [4,] Truthful TRUE FALSE # [five,] True True Imitation # [6,] False False Faux # ...
is.na(data) # x_num x_fac x_cha # [1,] Imitation Faux Simulated # [2,] Imitation Imitation TRUE # [3,] False Imitation Faux # [iv,] TRUE Truthful FALSE # [5,] Truthful Truthful FALSE # [6,] False FALSE FALSE # ...
The function produces a matrix, consisting of logical values (i.e. TRUE or FALSE), whereby TRUE indicates a missing value. Compare the output with the data table above — The True values are at the aforementioned position as before the NA elements.
An important characteristic of is.na is that the function can be reversed by but putting a ! (exclamation marker) in front end. In this case, TRUE indicates a value that is not NA in R:
! is . na (data) # x_num x_fac x_cha # [1,] True Truthful TRUE # [2,] TRUE Truthful FALSE # [3,] TRUE Truthful TRUE # [iv,] Fake FALSE True # [5,] FALSE Imitation True # [6,] True TRUE TRUE # ...
!is.na(data) # x_num x_fac x_cha # [1,] True True TRUE # [2,] TRUE TRUE Imitation # [iii,] Truthful True Truthful # [4,] Fake FALSE TRUE # [five,] Imitation Faux TRUE # [6,] TRUE Truthful True # ...
Exactly the reverse output equally before!
We are also able to cheque whether there is or is not an NA value in a column or vector:
is . na (data$x_num) # Works for numeric ... is . na (data$x_fac) # ... factor ... is . na (data$x_cha) # ... and character ! is . na (data$x_num) # The explanation marker still works ! is . na (information$x_fac) ! is . na (data$x_cha)
is.na(information$x_num) # Works for numeric ... is.na(data$x_fac) # ... gene ... is.na(data$x_cha) # ... and character !is.na(information$x_num) # The explanation mark still works !is.na(data$x_fac) !is.na(data$x_cha)
As you take seen, is.na provides u.s.a. with logical values that show us whether a value is NA or non. We tin can apply the role to a whole database or to a column (no matter which class the vector has).
That's nice, but the real power of is.na becomes visible in combination with other functions — And that's exactly what I'1000 going to show you at present.
On a side note:
R provides several other is.xxx functions that are very like to is.na (east.grand. is.nan, is.zilch, or is.finite). Stay tuned — All you learn here can be applied to many different programming scenarios!
is.na in Combination with Other R Functions
In the post-obit, I have prepared examples for the most important R functions that can be combined with is.na.
Remove NAs of Vector or Column
In a vector or column, NA values can exist removed as follows:
is . na_remove <- data$x_num[ ! is . na (data$x_num) ]
is.na_remove <- information$x_num[!is.na(information$x_num)]
Note: Our new vector is.na_remove is shorter in comparison to the original column information$x_num, since nosotros apply a filter that deletes all missing values.
Y'all can learn more about the removal of NA values from a vector here…
If you desire to drop rows with missing values of a information frame (i.due east. of multiple columns), the complete.cases role is preferable. Learn more…
Replace NAs with Other Values
Based on is.na, information technology is possible to replace NAs with other values such as cipher…
is . na_replace_0 <- data$x_num # Indistinguishable first cavalcade is . na_replace_0 [ is . na ( is . na_replace_0 ) ] <- 0 # Replace past 0
is.na_replace_0 <- data$x_num # Indistinguishable kickoff column is.na_replace_0[is.na(is.na_replace_0)] <- 0 # Replace past 0
…or the hateful.
is . na_replace_mean <- data$x_num # Duplicate first column x_num_mean <- hateful( is . na_replace_mean, na. rm = TRUE ) # Summate mean is . na_replace_mean [ is . na ( is . na_replace_mean ) ] <- x_num_mean # Replace past mean
is.na_replace_mean <- information$x_num # Duplicate first cavalcade x_num_mean <- mean(is.na_replace_mean, na.rm = TRUE) # Calculate hateful is.na_replace_mean[is.na(is.na_replace_mean)] <- x_num_mean # Replace by mean
In case of characters or factors, it is besides possible in R to set NA to bare:
is . na_blank_cha <- data$x_cha # Indistinguishable graphic symbol column is . na_blank_cha [ is . na ( is . na_blank_cha ) ] <- "" # Form graphic symbol to blank is . na_blank_fac <- data$x_fac # Duplicate factor column is . na_blank_fac <- equally . character ( is . na_blank_fac ) # Catechumen temporarily to character is . na_blank_fac [ is . na ( is . na_blank_fac ) ] <- "" # Course character to blank is . na_blank_fac <- every bit . cistron ( is . na_blank_fac ) # Recode dorsum to cistron
is.na_blank_cha <- data$x_cha # Duplicate graphic symbol column is.na_blank_cha[is.na(is.na_blank_cha)] <- "" # Class character to blank is.na_blank_fac <- information$x_fac # Duplicate gene column is.na_blank_fac <- every bit.grapheme(is.na_blank_fac) # Convert temporarily to character is.na_blank_fac[is.na(is.na_blank_fac)] <- "" # Class character to bare is.na_blank_fac <- every bit.factor(is.na_blank_fac) # Recode back to factor
Count NAs via sum & colSums
Combined with the R office sum, nosotros can count the corporeality of NAs in our columns. Co-ordinate to our previous data generation, it should exist approximately 20% in x_num, 30% in x_fac, and v% in x_cha.
sum( is . na (data$x_num) ) # 213 missings in the first column sum( is . na (data$x_fac) ) # 322 missings in the second column sum( is . na (information$x_cha) ) # 47 missings in the tertiary column
sum(is.na(data$x_num)) # 213 missings in the showtime column sum(is.na(data$x_fac)) # 322 missings in the second column sum(is.na(information$x_cha)) # 47 missings in the third column
If we want to count NAs in multiple columns at the same time, we can use the function colSums:
colSums( is . na (data) ) # x_num x_fac x_cha # 213 322 47
colSums(is.na(information)) # x_num x_fac x_cha # 213 322 47
Detect if there are any NAs
We tin also test, if there is at least 1 missing value in a cavalcade of our data. Equally we already know, it is TRUE that our columns have NAs.
any( is . na (data$x_num) ) # [ane] True
any(is.na(data$x_num)) # [1] Truthful
Locate NAs via which
In combination with the which function, is.na tin exist used to identify the positioning of NAs:
which( is . na (data$x_num) ) # [1] 4 five fourteen 17 22 23...
which(is.na(information$x_num)) # [1] four five 14 17 22 23...
Our outset column has missing values at the positions 4, v, 14, 17, 22, 23 and so along.
if & ifelse
Missing values have to be considered in our programming routines, e.m. within the if statement or within for loops.
In the following case, I'grand press "Damn, it'due south NA" to the R Studio panel whenever a missing occurs; and "Wow, that's awesome" in case of an observed value.
for (i in 1 :length(data$x_num) ) { if ( is . na (data$x_num[i] ) ) { print( "Damn, information technology's NA" ) } else { print( "Wow, that's awesome" ) } } # [1] "Wow, that's crawly" # [1] "Wow, that'south awesome" # [1] "Wow, that's awesome" # [1] "Damn, it'south NA" # [ane] "Damn, it'south NA" # [1] "Wow, that's crawly" # ...
for(i in 1:length(data$x_num)) { if(is.na(data$x_num[i])) { print("Damn, it's NA") } else { print("Wow, that's awesome") } } # [1] "Wow, that'southward awesome" # [ane] "Wow, that's awesome" # [1] "Wow, that's crawly" # [1] "Damn, it's NA" # [1] "Damn, information technology'south NA" # [ane] "Wow, that'southward crawly" # ...
Annotation: Within the if argument nosotros use is na instead of equal to — the approach we would usually use in case of observed values (e.grand. if(x[i] == 5)).
Fifty-fifty easier to apply: the ifelse function.
ifelse( is . na (data$x_num), "Damn, it'southward NA", "Wow, that'southward awesome" ) # [ane] "Wow, that's crawly" "Wow, that's awesome" "Wow, that's awesome" "Damn, it's NA" # [v] "Damn, it'due south NA" "Wow, that's awesome" ...
ifelse(is.na(data$x_num), "Damn, it's NA", "Wow, that'due south awesome") # [1] "Wow, that's awesome" "Wow, that'southward awesome" "Wow, that'due south awesome" "Damn, information technology's NA" # [v] "Damn, it'southward NA" "Wow, that's awesome" ...
Video Examples for the Handling of NAs in R
You lot want to learn even more than possibilities to deal with NAs in R? And then definitely bank check out the following video of my YouTuber channel.
In the video, I provide further examples for is.na. I besides speak nigh other functions for the handling of missing information in R data frames.
Now it'south on You!
I've shown yous the virtually important ways to use the is.na R office.
Even so, at that place are hundreds of different possibilities to apply is.na in a useful fashion.
Exercise you know whatsoever other helpful applications? Or do you have a question about the usage of is.na in a specific scenario?
Don't hesitate to permit me know in the comments!
Appendix
The header graphic of this page illustrates NA values in our data. The graphic can be produced with the post-obit R lawmaking:
N <- 2000 # Sample size ten <- runif(Due north) # Uniformly distributed variables y <- runif(Due north) x_NA <- runif( 50 ) # Random NAs y_NA <- runif( 50 ) par(bg = "#1b98e0" ) # Set background colour par(mar = c( 0, 0, 0, 0 ) ) # Remove space effectually plot pch_numb <- equally . character ( # Specify plotted numbers circular(runif(North, 0, 9 ) ) ) plot(ten, y, # Plot cex = ii, pch = pch_numb, col = "#353436" ) text(x_NA, y_NA, cex = two, # Add NA values to plot "NA", col = "red" ) points(10[ i : 500 ], y[ one : 500 ], # Overlay NA values with numbers cex = 2, pch = pch_numb, col = "#353436" )
N <- 2000 # Sample size x <- runif(N) # Uniformly distributed variables y <- runif(N) x_NA <- runif(50) # Random NAs y_NA <- runif(fifty) par(bg = "#1b98e0") # Set groundwork color par(mar = c(0, 0, 0, 0)) # Remove infinite around plot pch_numb <- every bit.character( # Specify plotted numbers round(runif(North, 0, nine))) plot(x, y, # Plot cex = 2, pch = pch_numb, col = "#353436") text(x_NA, y_NA, cex = 2, # Add NA values to plot "NA", col = "red") points(10[one:500], y[i:500], # Overlay NA values with numbers cex = ii, pch = pch_numb, col = "#353436")
Source: https://statisticsglobe.com/r-is-na-function/

0 Response to "How To Filter Out Na In R"
Post a Comment